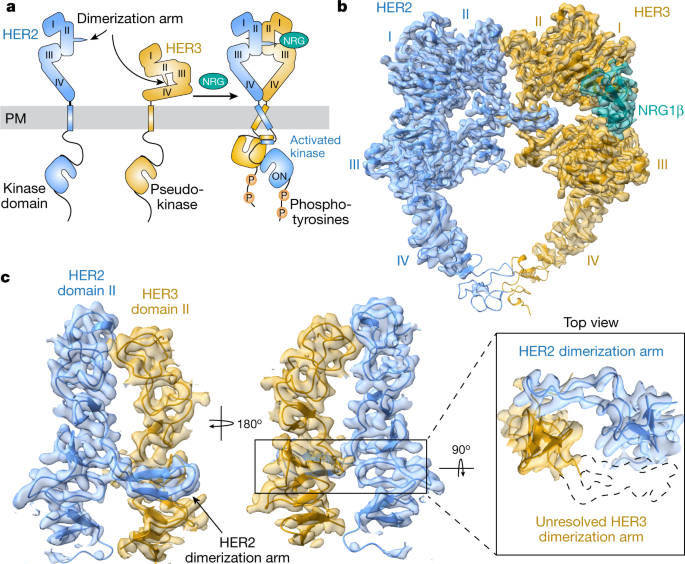
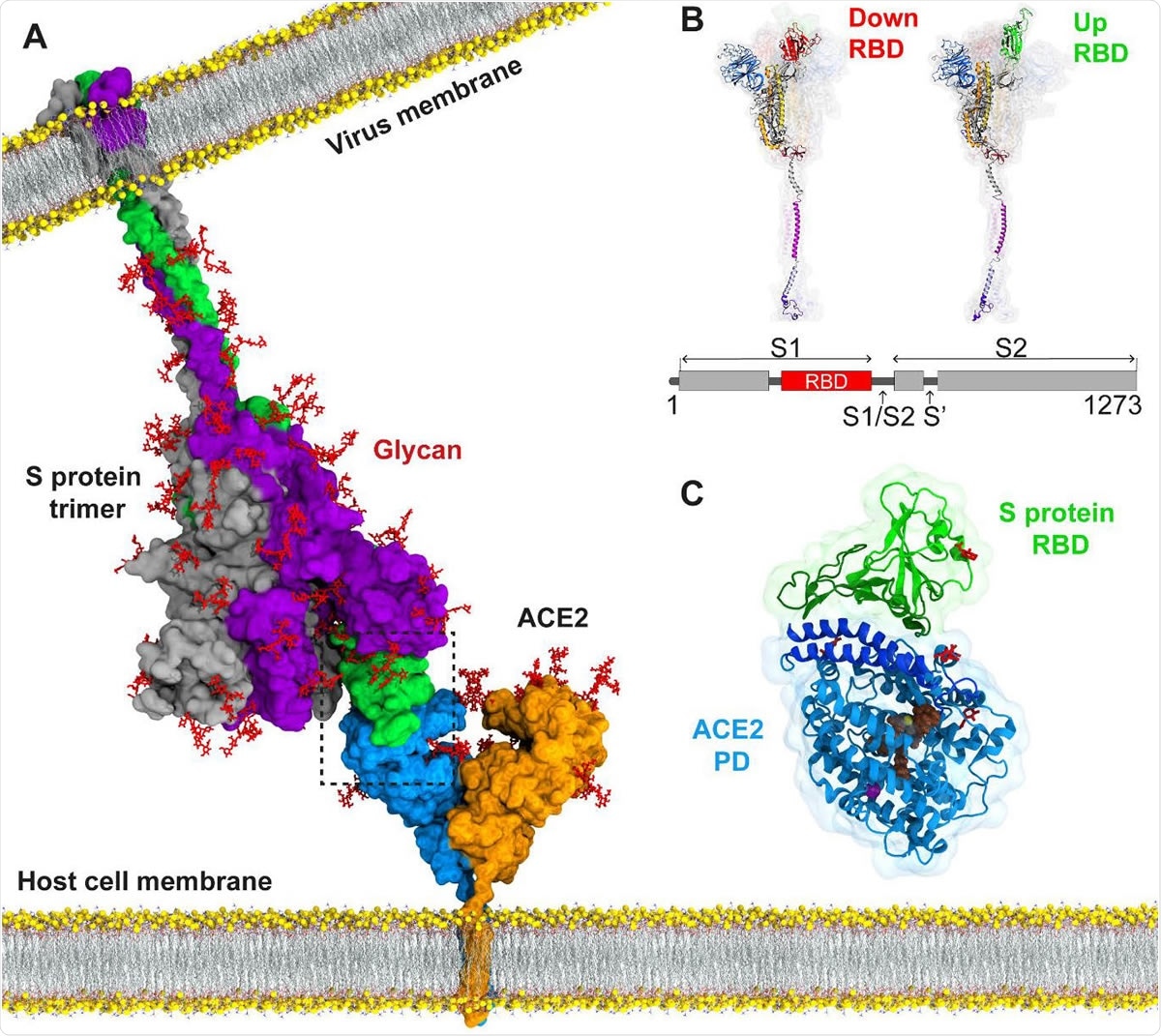
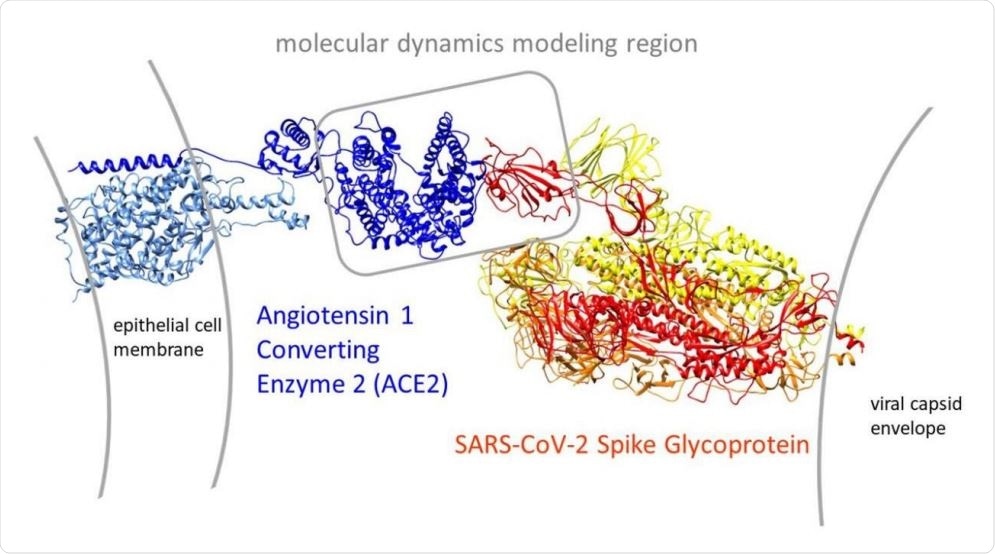
Molecular dynamic simulations reveal detailed spike-ACE2 interactions
4.9 (469) · $ 25.50 · In stock
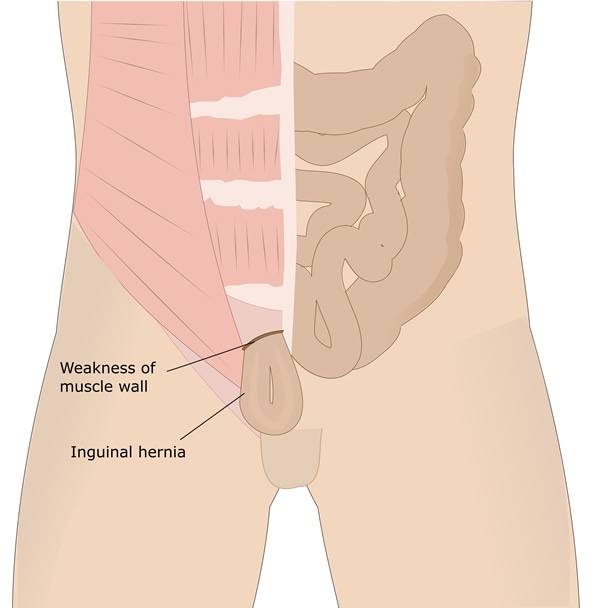
The current COVID-19 pandemic has spread throughout the world. Caused by a single-stranded RNA betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is closely related to but much more infectious than the earlier highly pathogenic betacoronaviruses SARS and MERS-CoV, has impacted social, economic, and physical health to an unimaginable extent.

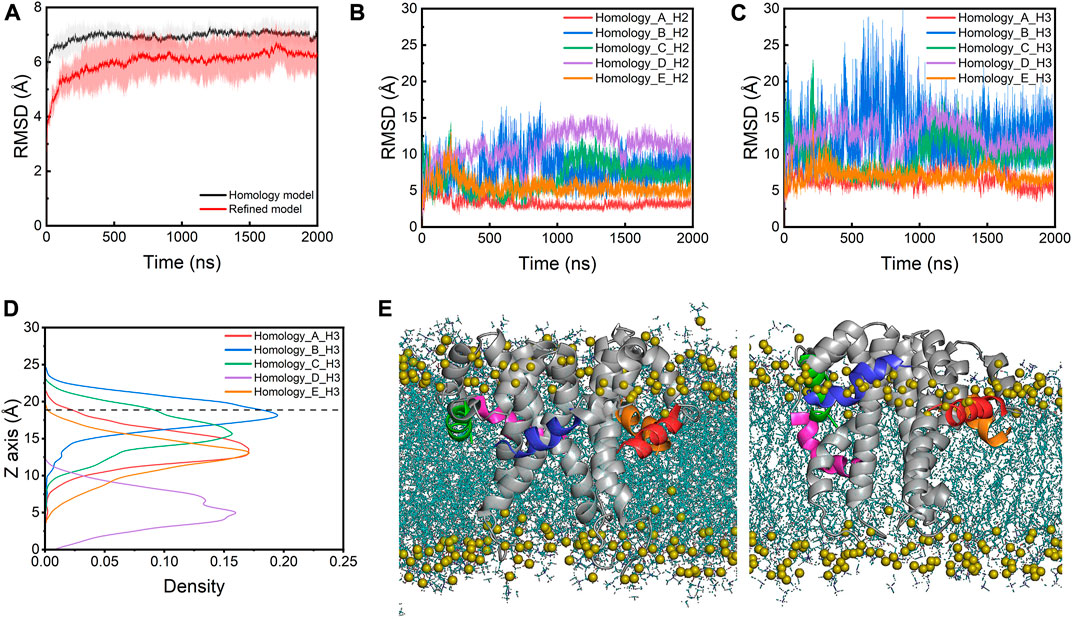
The molecular dynamic simulation results of interaction between
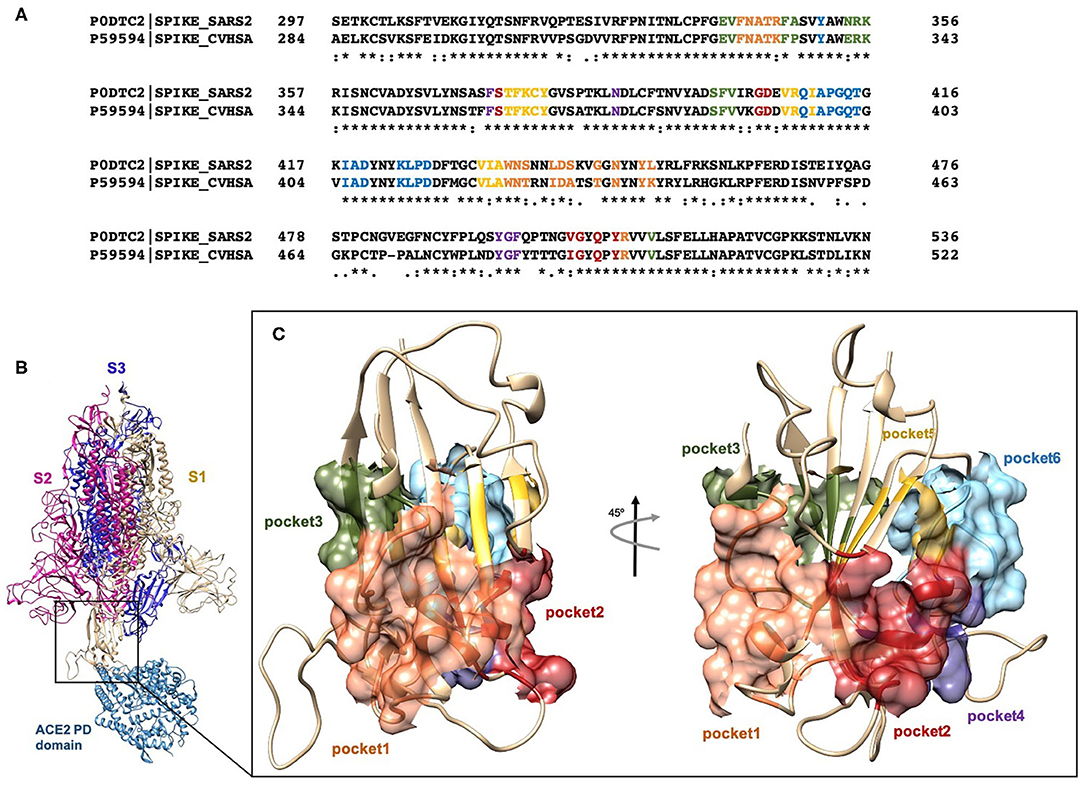
Frontiers Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain

Raman spectroscopy study of 7,8-dihydrofolate inhibition on the Wuhan strain SARS-CoV-2 binding to human ACE2 receptor - ScienceDirect
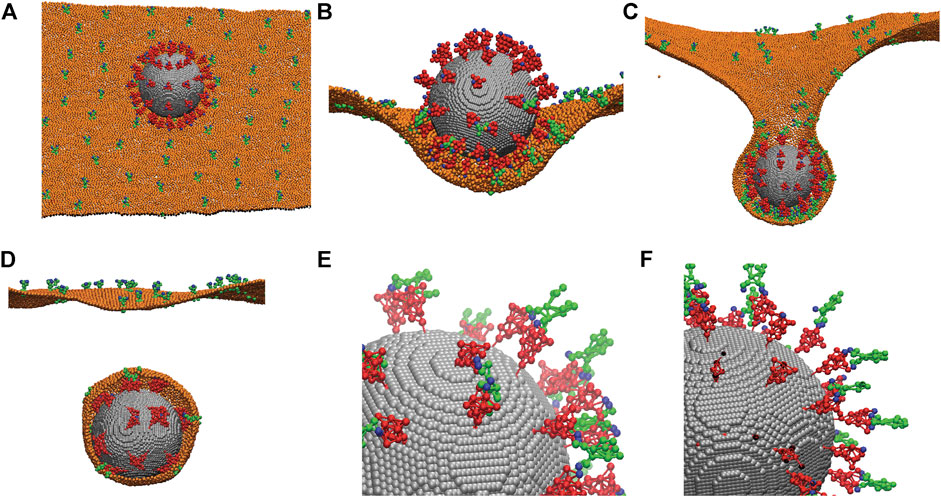
Frontiers Refinement of SARS-CoV-2 envelope protein structure in a native-like environment by molecular dynamics simulations
Computational models screen for coronaviruses likely to infect humans
Frontiers Coarse-Grained Modeling of Coronavirus Spike Proteins and ACE2 Receptors
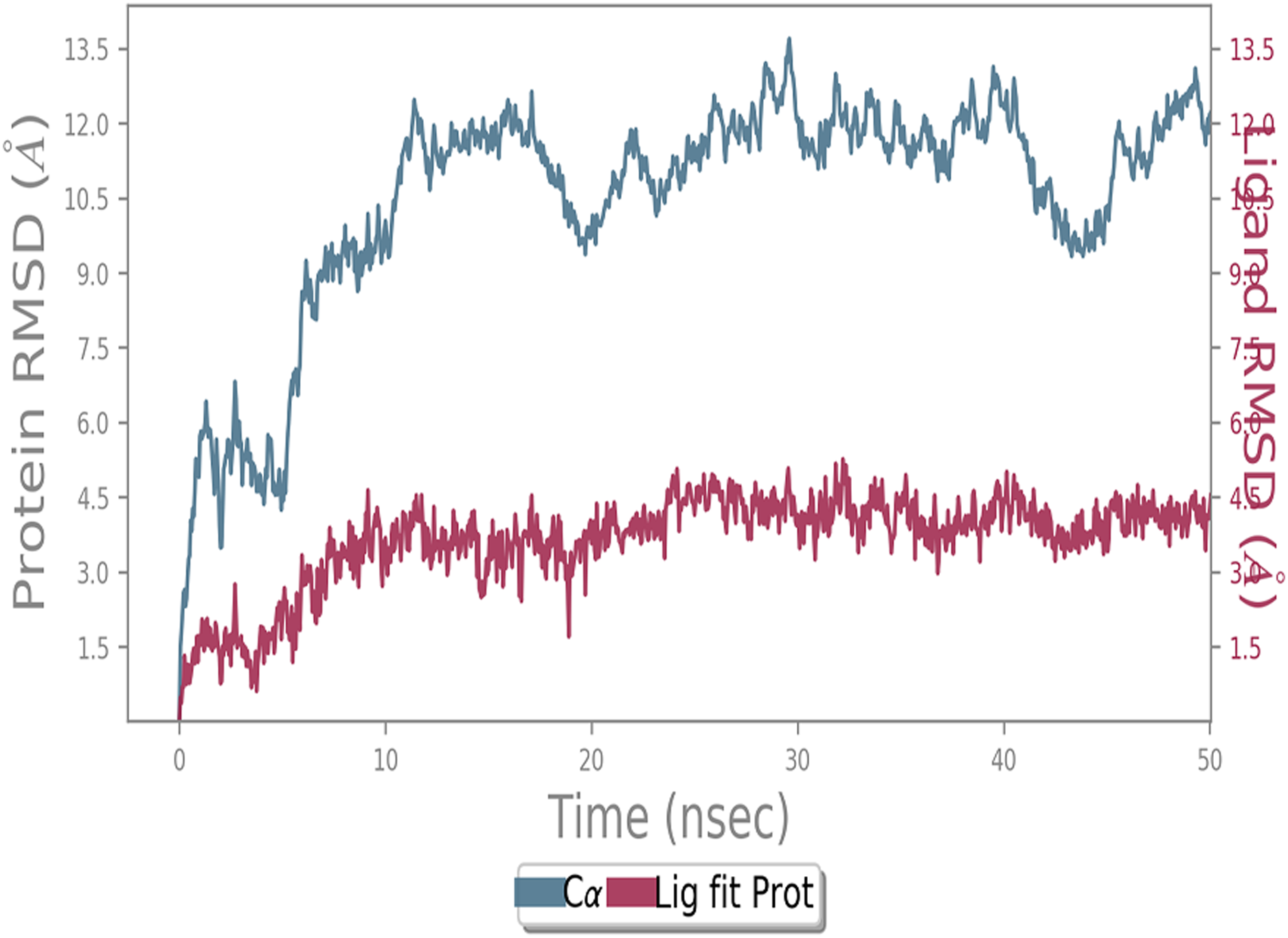
A molecular dynamics simulation study of the ACE2 receptor with screened natural inhibitors to identify novel drug candidate against COVID-19 [PeerJ]

Self-Association of ACE-2 with Different RBD Amounts: A Dynamic Simulation Perspective on SARS-CoV-2 Infection

Molecular Interaction And Inhibition Of SARS-CoV-2 Binding, 54% OFF
Ai helps identify critical interactions for SARS-CoV-2 spike protein binding to ACE2

SARS-CoV-2 spike binding to ACE2 is stronger and longer ranged due to glycan interaction - ScienceDirect

Full article: Virtual screening and molecular dynamics simulation study of abyssomicins as potential inhibitors of COVID‐19 virus main protease and spike protein